
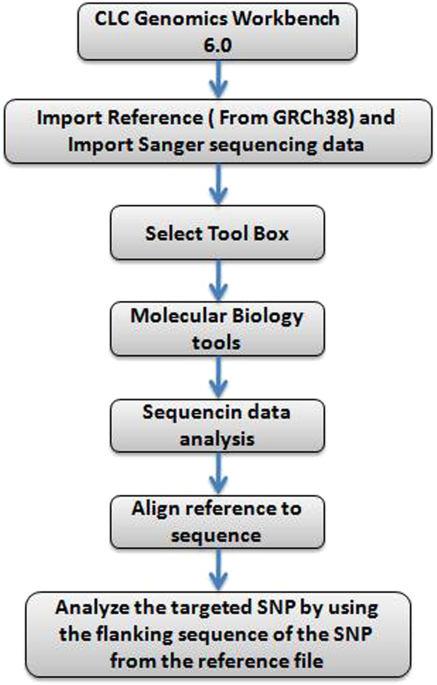
We sequenced ~1% of the orangutan genome with 41-fold median coverage in 31 wild-born individuals from two populations. Our modifications facilitate generation of single-sample libraries, enabling individual genotype assignments instead of pooled-sample analysis.
From the bioinformatical perspective, the reliance of most studies on a single SNP caller disregards the possibility that different algorithms may produce disparate SNP datasets.Results: We present an improved RRL (iRRL) protocol that maximizes the generation of homologous DNA sequences, thus achieving improved genotyping-by-sequencing efficiency.
In the laboratory, current protocols require improvements with regards to sequencing homologous fragments to reduce the number of missing genotypes. Yet, generating such datasets remains challenging due to laboratory and bioinformatical issues. Like similar approaches, RRL sequencing reduces ascertainment bias due to simultaneous discovery and genotyping of single-nucleotide polymorphisms (SNPs) and does not require reference genomes. investigating only parts of the genome, is reduced-representation library (RRL) sequencing. One approach to reduce genome complexity, i.e. Available under: doi: 10.1186/1471- BibTex of SNP datasets for orangutan population genomics using improved reduced-representation sequencing and direct comparisons of SNP calling algorithms},Īuthor=,īackground: High-throughput sequencing has opened up exciting possibilities in population and conservation genetics by enabling the assessment of genetic variation at genome-wide scales. Generation of SNP datasets for orangutan population genomics using improved reduced-representation sequencing and direct comparisons of SNP calling algorithms. STÖLTING, Alexander NATER, Benoit GOOSSENS, Natasha ARORA, Rémy BRUGGMANN, Andrea PATRIGNANI, Beatrice NUSSBERGER, Reeta SHARMA, Robert KRAUS, 2014. This study presents the first effort to generate a population genomic dataset for wild-born orangutans with known population provenance. These differences affected scans for signatures of natural selection, but will also exert undue influences on demographic inferences. Our direct comparison of three commonly used SNP callers emphasizes the need to question the accuracy of SNP and genotype calling, as we obtained considerably different SNP datasets depending on caller algorithms, sequencing depths and filtering criteria. Furthermore, conflicting genotypes between two algorithms showed a systematic bias in that one caller almost exclusively assigned heterozygotes, while the other one almost exclusively assigned homozygotes.Ĭonclusions: Our enhanced iRRL approach greatly facilitates genotyping-by-sequencing and thus direct estimates of allele frequencies. Of all conflicting genotype calls, CLC was only correct in 17% of the cases. Genotype validations revealed that the Unified Genotyper of the Genome Analysis Toolkit and SAMtools performed significantly better than a caller from CLC Genomics Workbench (CLC). We obtained substantially different SNP datasets depending on the SNP caller. SNPs and genotypes were called using three different algorithms. Results: We present an improved RRL (iRRL) protocol that maximizes the generation of homologous DNA sequences, thus achieving improved genotyping-by-sequencing efficiency. From the bioinformatical perspective, the reliance of most studies on a single SNP caller disregards the possibility that different algorithms may produce disparate SNP datasets. Background: High-throughput sequencing has opened up exciting possibilities in population and conservation genetics by enabling the assessment of genetic variation at genome-wide scales.


 0 kommentar(er)
0 kommentar(er)
